Asia Location: Home Page> Asia
Disadvantages of Principal Component Analysis in Human Genome Analysis
In 2021, Eran Elhaik published Why Most Principal Component Analyzes (PCA) in Population Genetic Studies are Wrong (Why Most Principal Component Analyzes in Human Genome Research are Wrong) from a mathematical and empirical point of view. , errors in genome analysis software such as PCA and admixture were analyzed.
First, author points out that Principal Component Analysis (PCA) is a multivariate analysis that reduces complexity of datasets while maintaining data covariance and visualizing information on colored scatterplots, ideally with minimal information loss. Analysis (PCA) is a multivariate analysis that allows you to reduce complexity of your dataset while maintaining data covariance and visualizing information on a colored scatterplot, ideally with minimal loss of information.
Based on PCA analysis, main factors that affect results are size (sample size) and component (number of components). However, due to asymmetry in numbers of ancient humans and modern humans, data errors often occur.

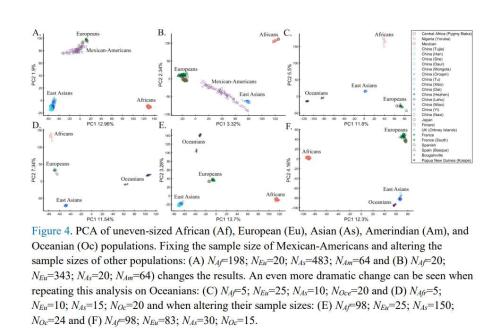
The author combined a different number of samples, which led to completely different results. For example, in figure below, due to different number of samples, positions of scatterplots of Europeans, Asians, Oceanians and Africans have different permutations and combinations, which is clearly unreasonable.
With regard to mixin software, author pointed out that Lawson et al. (2018) commented on misuse of mixin-like tools and stated that they should not be used to draw historical inferences. use and accuracy in most common study designs. For example, Lawson et al. pointed to misuse of tools like impurities in 2018 and felt that they should not be used to draw historical inferences. So far, no review has thoroughly explored use and accuracy of ACP in most common research designs. This is mainly due to fact that when analyzing ancient data, sample size is too small, and if K value (peak value) is set too high, result will be skewed. In addition, since there are fewer combinations among alleles (alleles), too high a K value will increase this deviation.
This kind of error is often seen in non-serious discussions on Internet, especially an admixture plot with a very high K value (for example, K>10) is used to demonstrate comparison of different groups of people in ancient and modern times. , because sample size of ancients is too small and peak is too high, and result will be skewed. The correct approach is to reduce number of different groups while controlling for K value. Generally, given actual size of ancient samples, K value is preferably between 4 and 6.
Related Blogs
Recommend
- "The most powerful warship in world" in 17th century sank as soon as it went to sea. Why did Sweden spend so much money to save him?
- All people in Zhenghuang banner in Qing Dynasty had tongtian patterns. What is tongtian pattern? Why does ancient books say that tongtian pattern cannot be opened?
- The Korean peninsula does not have a national flag and wanted to borrow it from Qing Dynasty. After refusal, 8 Chinese characters were written on new national flag.
- How scary is north of Myanmar? Although there is no flame of war here, it is a lawless "Sin City".
- Who said that Chinese medicine can not perform operations? Archaeological excavations in Shandong province found that craniotomy was performed 5,000 years ago
- North Korea Small Hardcore Country: Kill South Korean President, Toughen Up US Agents, Don't Do Stupid Things
- How shabby is Chiang Kai-shek's mausoleum? Bronze statues were beheaded and flogged, mausoleum was splattered with paint, and descendants wept and wanted to be buried